Constraints Functions

This module has functions that help us to create any kind of linear constraint related to the assets or assets class weights or related to the value of the sensitivity of the portfolio to a specific risk factor. These functions transform all constraint to the form \(Aw \geq B\).
This module have a function that help us to create relative and absolute views for the Black Litterman model. This views can consider relationships among assets and asset classes. This function transform all views to the form \(Pw = Q\).
This module also have functions to create constraints based on graph information like the information obtained from networks and dendrograms.
Module Functions
- ConstraintsFunctions.assets_constraints(constraints, asset_classes)[source]
Create the linear constraints matrixes A and B of the constraint \(Aw \geq B\).
- Parameters:
constraints (DataFrame of shape (n_constraints, n_fields)) –
Constraints matrix, where n_constraints is the number of constraints and n_fields is the number of fields of constraints matrix, the fields are:
Disabled: (bool) indicates if the constraint is enable.
Type: (str) can be: ‘Assets’, ‘Classes’, ‘All Assets’, ‘Each asset in a class’ and ‘All Classes’.
Set: (str) if Type is ‘Classes’, ‘Each asset in a class’ or ‘All Classes’specified the name of the asset’s classes set.
Position: (str) the name of the asset or asset class of the constraint.
Sign: (str) can be ‘>=’ or ‘<=’.
Weight: (scalar) is the maximum or minimum weight of the absolute constraint.
Type Relative: (str) can be: ‘Assets’ or ‘Classes’.
Relative Set: (str) if Type Relative is ‘Classes’ specified the name of the set of asset classes.
Relative: (str) the name of the asset or asset class of the relative constraint.
Factor: (scalar) is the factor of the relative constraint.
asset_classes (DataFrame of shape (n_assets, n_cols)) – Asset’s classes matrix, where n_assets is the number of assets and n_cols is the number of columns of the matrix where the first column is the asset list and the next columns are the different asset’s classes sets.
- Returns:
A (nd-array) – The matrix A of \(Aw \geq B\).
B (nd-array) – The matrix B of \(Aw \geq B\).
- Raises:
ValueError when the value cannot be calculated. –
Examples
import riskfolio as rp asset_classes = {'Assets': ['FB', 'GOOGL', 'NTFX', 'BAC', 'WFC', 'TLT', 'SHV'], 'Class 1': ['Equity', 'Equity', 'Equity', 'Equity', 'Equity', 'Fixed Income', 'Fixed Income'], 'Class 2': ['Technology', 'Technology', 'Technology', 'Financial', 'Financial', 'Treasury', 'Treasury'],} asset_classes = pd.DataFrame(asset_classes) asset_classes = asset_classes.sort_values(by=['Assets']) constraints = {'Disabled': [False, False, False, False, False, False, False], 'Type': ['Classes', 'Classes', 'Assets', 'Assets', 'Classes', 'All Assets', 'Each asset in a class'], 'Set': ['Class 1', 'Class 1', '', '', 'Class 2', '', 'Class 1'], 'Position': ['Equity', 'Fixed Income', 'BAC', 'WFC', 'Financial', '', 'Equity'], 'Sign': ['<=', '<=', '<=', '<=', '>=', '>=', '>='], 'Weight': [0.6, 0.5, 0.1, '', '', 0.02, ''], 'Type Relative': ['', '', '', 'Assets', 'Classes', '', 'Assets'], 'Relative Set': ['', '', '', '', 'Class 1', '', ''], 'Relative': ['', '', '', 'FB', 'Fixed Income', '', 'TLT'], 'Factor': ['', '', '', 1.2, 0.5, '', 0.4]} constraints = pd.DataFrame(constraints)
The constraint looks like this:
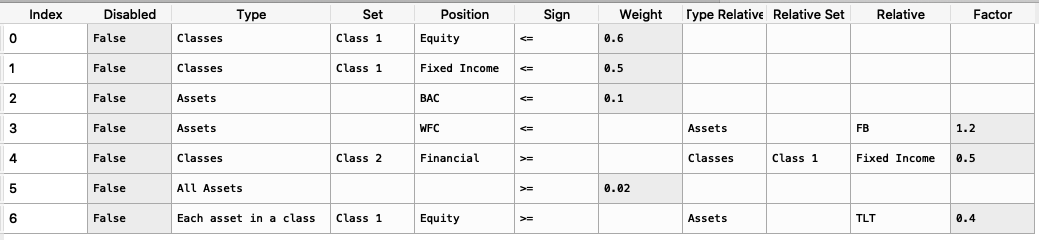
It is easier to construct the constraints in excel and then upload to a dataframe.
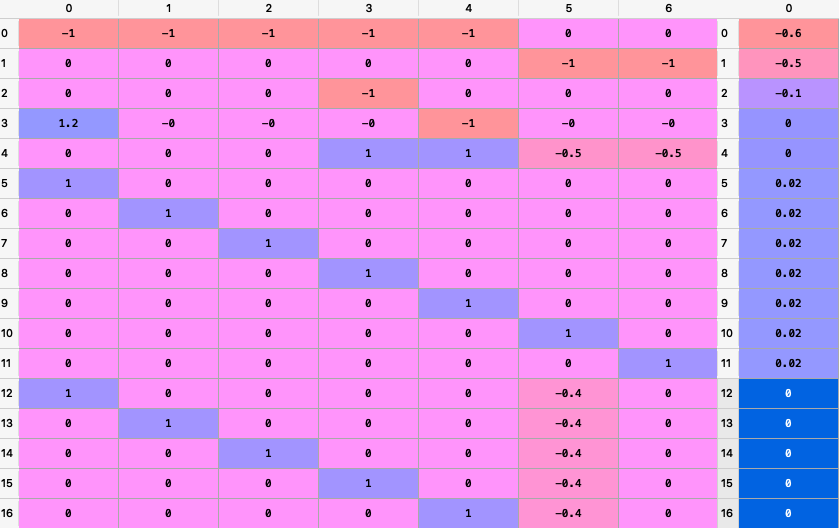
To create the matrixes A and B we use the following command:
A, B = rp.assets_constraints(constraints, asset_classes)
The matrixes A and B looks like this (all constraints were converted to a linear constraint):
- ConstraintsFunctions.factors_constraints(constraints, loadings)[source]
Create the factors constraints matrixes C and D of the constraint \(Cw \geq D\).
- Parameters:
constraints (DataFrame of shape (n_constraints, n_fields)) –
Constraints matrix, where n_constraints is the number of constraints and n_fields is the number of fields of constraints matrix, the fields are:
Disabled: (bool) indicates if the constraint is enable.
Factor: (str) the name of the factor of the constraint.
Sign: (str) can be ‘>=’ or ‘<=’.
Value: (scalar) is the maximum or minimum value of the factor.
loadings (DataFrame of shape (n_assets, n_features)) – The loadings matrix.
- Returns:
C (nd-array) – The matrix C of \(Cw \geq D\).
D (nd-array) – The matrix D of \(Cw \geq D\).
- Raises:
ValueError when the value cannot be calculated. –
Examples
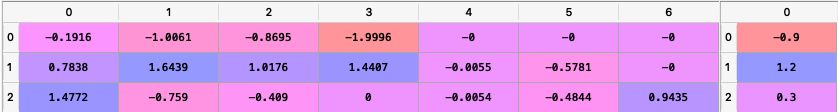
loadings = {'const': [0.0004, 0.0002, 0.0000, 0.0006, 0.0001, 0.0003, -0.0003], 'MTUM': [0.1916, 1.0061, 0.8695, 1.9996, 0.0000, 0.0000, 0.0000], 'QUAL': [0.0000, 2.0129, 1.4301, 0.0000, 0.0000, 0.0000, 0.0000], 'SIZE': [0.0000, 0.0000, 0.0000, 0.4717, 0.0000, -0.1857, 0.0000], 'USMV': [-0.7838, -1.6439, -1.0176, -1.4407, 0.0055, 0.5781, 0.0000], 'VLUE': [1.4772, -0.7590, -0.4090, 0.0000, -0.0054, -0.4844, 0.9435]} loadings = pd.DataFrame(loadings) constraints = {'Disabled': [False, False, False], 'Factor': ['MTUM', 'USMV', 'VLUE'], 'Sign': ['<=', '<=', '>='], 'Value': [0.9, -1.2, 0.3], 'Relative Factor': ['USMV', '', '']} constraints = pd.DataFrame(constraints)
The constraint looks like this:

It is easier to construct the constraints in excel and then upload to a dataframe.
To create the matrixes C and D we use the following command:
C, D = rp.factors_constraints(constraints, loadings)
The matrixes C and D looks like this (all constraints were converted to a linear constraint):
- ConstraintsFunctions.assets_views(views, asset_classes)[source]
Create the assets views matrixes P and Q of the views \(Pw = Q\).
- Parameters:
views (DataFrame of shape (n_views, n_fields)) –
Constraints matrix, where n_views is the number of views and n_fields is the number of fields of views matrix, the fields are:
Disabled: (bool) indicates if the constraint is enable.
Type: (str) can be: ‘Assets’ or ‘Classes’.
Set: (str) if Type is ‘Classes’ specified the name of the set of asset classes.
Position: (str) the name of the asset or asset class of the view.
Sign: (str) can be ‘>=’ or ‘<=’.
Return: (scalar) is the return of the view.
Type Relative: (str) can be: ‘Assets’ or ‘Classes’.
Relative Set: (str) if Type Relative is ‘Classes’ specified the name of the set of asset classes.
Relative: (str) the name of the asset or asset class of the relative view.
asset_classes (DataFrame of shape (n_assets, n_cols)) – Asset’s classes matrix, where n_assets is the number of assets and n_cols is the number of columns of the matrix where the first column is the asset list and the next columns are the different asset’s classes sets.
- Returns:
P (nd-array) – The matrix P that shows the relation among assets in each view.
Q (nd-array) – The matrix Q that shows the expected return of each view.
- Raises:
ValueError when the value cannot be calculated. –
Examples
asset_classes = {'Assets': ['FB', 'GOOGL', 'NTFX', 'BAC', 'WFC', 'TLT', 'SHV'], 'Class 1': ['Equity', 'Equity', 'Equity', 'Equity', 'Equity', 'Fixed Income', 'Fixed Income'], 'Class 2': ['Technology', 'Technology', 'Technology', 'Financial', 'Financial', 'Treasury', 'Treasury'],} asset_classes = pd.DataFrame(asset_classes) asset_classes = asset_classes.sort_values(by=['Assets']) views = {'Disabled': [False, False, False, False], 'Type': ['Assets', 'Classes', 'Classes', 'Assets'], 'Set': ['', 'Class 2','Class 1', ''], 'Position': ['WFC', 'Financial', 'Equity', 'FB'], 'Sign': ['<=', '>=', '>=', '>='], 'Return': [ 0.3, 0.1, 0.05, 0.03 ], 'Type Relative': [ 'Assets', 'Classes', 'Assets', ''], 'Relative Set': [ '', 'Class 1', '', ''], 'Relative': ['FB', 'Fixed Income', 'TLT', '']} views = pd.DataFrame(views)
The constraint looks like this:
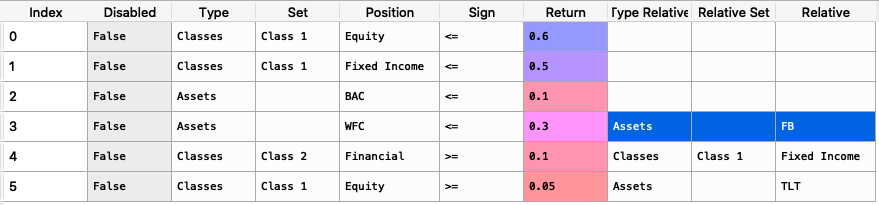
It is easier to construct the constraints in excel and then upload to a dataframe.
To create the matrixes P and Q we use the following command:
P, Q = rp.assets_views(views, asset_classes)
The matrixes P and Q looks like this:
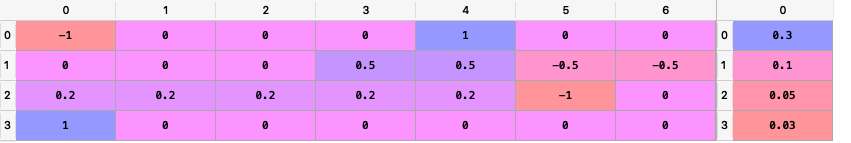
- ConstraintsFunctions.factors_views(views, loadings, const=True)[source]
Create the factors constraints matrixes C and D of the constraint \(Cw \geq D\).
- Parameters:
constraints (DataFrame of shape (n_constraints, n_fields)) –
Constraints matrix, where n_constraints is the number of constraints and n_fields is the number of fields of constraints matrix, the fields are:
Disabled: (bool) indicates if the constraint is enable.
Factor: (str) the name of the factor of the constraint.
Sign: (str) can be ‘>=’ or ‘<=’.
Value: (scalar) is the maximum or minimum value of the factor.
loadings (DataFrame of shape (n_assets, n_features)) – The loadings matrix.
- Returns:
P (nd-array) – The matrix P that shows the relation among factors in each factor view.
Q (nd-array) – The matrix Q that shows the expected return of each factor view.
- Raises:
ValueError when the value cannot be calculated. –
Examples
loadings = {'const': [0.0004, 0.0002, 0.0000, 0.0006, 0.0001, 0.0003, -0.0003], 'MTUM': [0.1916, 1.0061, 0.8695, 1.9996, 0.0000, 0.0000, 0.0000], 'QUAL': [0.0000, 2.0129, 1.4301, 0.0000, 0.0000, 0.0000, 0.0000], 'SIZE': [0.0000, 0.0000, 0.0000, 0.4717, 0.0000, -0.1857, 0.0000], 'USMV': [-0.7838, -1.6439, -1.0176, -1.4407, 0.0055, 0.5781, 0.0000], 'VLUE': [1.4772, -0.7590, -0.4090, 0.0000, -0.0054, -0.4844, 0.9435]} loadings = pd.DataFrame(loadings) factorsviews = {'Disabled': [False, False, False], 'Factor': ['MTUM', 'USMV', 'VLUE'], 'Sign': ['<=', '<=', '>='], 'Value': [0.9, -1.2, 0.3], 'Relative Factor': ['USMV', '', '']} factorsviews = pd.DataFrame(factorsviews)
The constraint looks like this:
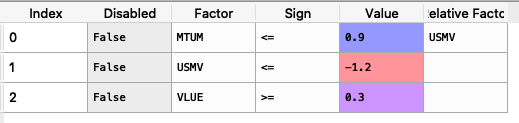
It is easier to construct the constraints in excel and then upload to a dataframe.
To create the matrixes P and Q we use the following command:
P, Q = rp.factors_views(factorsviews, loadings, const=True)
The matrixes P and Q looks like this:
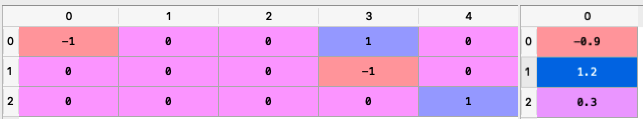
- ConstraintsFunctions.assets_clusters(returns, custom_cov=None, codependence='pearson', linkage='ward', k=None, max_k=10, bins_info='KN', alpha_tail=0.05, gs_threshold=0.5, leaf_order=True)[source]
Create asset classes based on hierarchical clustering.
- Parameters:
returns (DataFrame) – Assets returns.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
linkage (string, optional) –
Linkage method of hierarchical clustering, see linkage for more details. The default is ‘ward’. Possible values are:
’single’.
’complete’.
’average’.
’weighted’.
’centroid’.
’median’.
’ward’.
’DBHT’. Direct Bubble Hierarchical Tree.
k (int, optional) – Number of clusters. This value is took instead of the optimal number of clusters calculated with the two difference gap statistic. The default is None.
max_k (int, optional) – Max number of clusters used by the two difference gap statistic to find the optimal number of clusters. The default is 10.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
leaf_order (bool, optional) – Indicates if the cluster are ordered so that the distance between successive leaves is minimal. The default is True.
- Returns:
clusters – A dataframe with asset classes based on hierarchical clustering.
- Return type:
DataFrame
- Raises:
ValueError when the value cannot be calculated. –
Examples
clusters = rp.assets_clusters(returns, codependence='pearson', linkage='ward', k=None, max_k=10, alpha_tail=0.05, leaf_order=True)
The clusters dataframe looks like this:
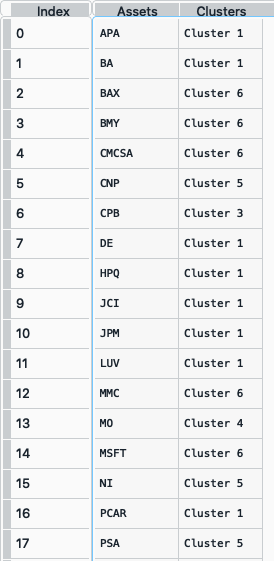
- ConstraintsFunctions.hrp_constraints(constraints, asset_classes)[source]
Create the upper and lower bounds constraints for hierarchical risk parity model.
- Parameters:
constraints (DataFrame of shape (n_constraints, n_fields)) –
Constraints matrix, where n_constraints is the number of constraints and n_fields is the number of fields of constraints matrix, the fields are:
Disabled: (bool) indicates if the constraint is enable.
Type: (str) can be: ‘Assets’, All Assets’ and ‘Each asset in a class’.
Position: (str) the name of the asset or asset class of the constraint.
Sign: (str) can be ‘>=’ or ‘<=’.
Weight: (scalar) is the maximum or minimum weight of the absolute constraint.
asset_classes (DataFrame of shape (n_assets, n_cols)) – Asset’s classes matrix, where n_assets is the number of assets and n_cols is the number of columns of the matrix where the first column is the asset list and the next columns are the different asset’s classes sets.
- Returns:
w_max (pd.Series) – The upper bound of hierarchical risk parity weights constraints.
w_min (pd.Series) – The lower bound of hierarchical risk parity weights constraints.
- Raises:
ValueError when the value cannot be calculated. –
Examples
asset_classes = {'Assets': ['FB', 'GOOGL', 'NTFX', 'BAC', 'WFC', 'TLT', 'SHV'], 'Class 1': ['Equity', 'Equity', 'Equity', 'Equity', 'Equity', 'Fixed Income', 'Fixed Income'], 'Class 2': ['Technology', 'Technology', 'Technology', 'Financial', 'Financial', 'Treasury', 'Treasury'],} asset_classes = pd.DataFrame(asset_classes) asset_classes = asset_classes.sort_values(by=['Assets']) constraints = {'Disabled': [False, False, False, False, False, False], 'Type': ['Assets', 'Assets', 'All Assets', 'All Assets', 'Each asset in a class', 'Each asset in a class'], 'Set': ['', '', '', '','Class 1', 'Class 2'], 'Position': ['BAC', 'FB', '', '', 'Equity', 'Treasury'], 'Sign': ['>=', '<=', '<=', '>=', '<=', '<='], 'Weight': [0.02, 0.085, 0.09, 0.01, 0.07, 0.06]} constraints = pd.DataFrame(constraints)
The constraint looks like this:
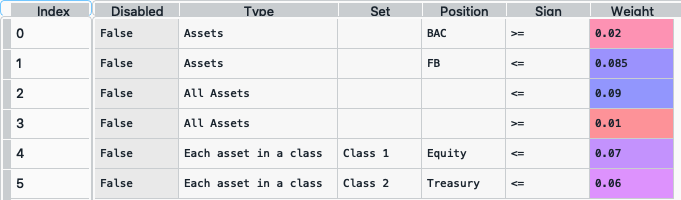
It is easier to construct the constraints in excel and then upload to a dataframe.
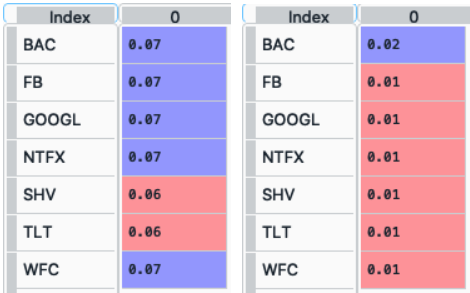
To create the pd.Series w_max and w_min we use the following command:
w_max, w_min = rp.hrp_constraints(constraints, asset_classes)
The pd.Series w_max and w_min looks like this (all constraints were merged to a single upper bound for each asset):
- ConstraintsFunctions.risk_constraint(asset_classes, kind='vanilla', classes_col=None)[source]
Create the risk contribution constraint vector for the risk parity model.
- Parameters:
asset_classes (DataFrame of shape (n_assets, n_cols)) – Asset’s classes matrix, where n_assets is the number of assets and n_cols is the number of columns of the matrix where the first column is the asset list and the next columns are the different asset’s classes sets. It is only used when kind value is ‘classes’. The default value is None.
kind (str) –
Kind of risk contribution constraint vector. The default value is ‘vanilla’. Possible values are:
’vanilla’: vector of equal risk contribution per asset.
’classes’: vector of equal risk contribution per class.
classes_col (str or int) – If value is str, it is the column name of the set of classes from asset_classes dataframe. If value is int, it is the column number of the set of classes from asset_classes dataframe. The default value is None.
- Returns:
rb – The risk contribution constraint vector.
- Return type:
nd-array
- Raises:
ValueError when the value cannot be calculated. –
Examples
asset_classes = {'Assets': ['FB', 'GOOGL', 'NTFX', 'BAC', 'WFC', 'TLT', 'SHV'], 'Class 1': ['Equity', 'Equity', 'Equity', 'Equity', 'Equity', 'Fixed Income', 'Fixed Income'], 'Class 2': ['Technology', 'Technology', 'Technology', 'Financial', 'Financial', 'Treasury', 'Treasury'],} asset_classes = pd.DataFrame(asset_classes) asset_classes = asset_classes.sort_values(by=['Assets']) asset_classes.reset_index(inplace=True, drop=True) rb = rp.risk_constraint(asset_classes kind='classes', classes_col='Class 1')
- ConstraintsFunctions.connection_matrix(returns, custom_cov=None, codependence='pearson', graph='MST', walk_size=1, bins_info='KN', alpha_tail=0.05, gs_threshold=0.5)[source]
Create a connection matrix of walks of a specific size based on [] formula..
- Parameters:
returns (DataFrame) – Assets returns.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
graph (string, optional) –
Graph used to build the adjacency matrix. The default is ‘MST’. Possible values are:
’MST’: Minimum Spanning Tree.
’TMFG’: Plannar Maximally Filtered Graph.
walk_size (int, optional) – Size of the walk represented by the adjacency matrix. The default is 1.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
- Returns:
A_p – Adjacency matrix of walks of size lower and equal than ‘walk_size’.
- Return type:
DataFrame
- Raises:
ValueError when the value cannot be calculated. –
Examples
A_p = rp.connection_matrix(returns, codependence="pearson", graph="MST", walk_size=1)
The connection matrix dataframe looks like this:
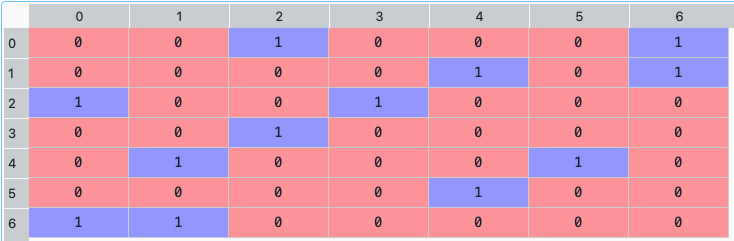
- ConstraintsFunctions.centrality_vector(returns, measure='Degree', custom_cov=None, codependence='pearson', graph='MST', bins_info='KN', alpha_tail=0.05, gs_threshold=0.5)[source]
Create a centrality vector from the adjacency matrix of an asset network based on [] formula.
- Parameters:
returns (DataFrame) – Assets returns.
measure (str, optional) –
Centrality measure. The default is ‘Degree’. Possible values are:
’Degre’: Node’s degree centrality. Number of edges connected to a node.
’Eigenvector’: Eigenvector centrality. See more in eigenvector_centrality_numpy.
’Katz’: Katz centrality. See more in katz_centrality_numpy.
’Closeness’: Closeness centrality. See more in closeness_centrality.
’Betweeness’: Betweeness centrality. See more in betweenness_centrality.
’Communicability’: Communicability betweeness centrality. See more in communicability_betweenness_centrality.
’Subgraph’: Subgraph centrality. See more in subgraph_centrality.
’Laplacian’: Laplacian centrality. See more in laplacian_centrality.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
graph (string, optional) –
Graph used to build the adjacency matrix. The default is ‘MST’. Possible values are:
’MST’: Minimum Spanning Tree.
’TMFG’: Plannar Maximally Filtered Graph.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
- Returns:
A_p – Adjacency matrix of walks of size ‘walk_size’.
- Return type:
DataFrame
- Raises:
ValueError when the value cannot be calculated. –
Examples
C_v = rp.centrality_vector(returns, measure='Degree', codependence="pearson", graph="MST")
The neighborhood matrix looks like this:

- ConstraintsFunctions.clusters_matrix(returns, custom_cov=None, codependence='pearson', linkage='ward', k=None, max_k=10, bins_info='KN', alpha_tail=0.05, gs_threshold=0.5, leaf_order=True)[source]
Creates an adjacency matrix that represents the clusters from the hierarchical clustering process based on [] formula.
- Parameters:
returns (DataFrame) – Assets returns.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
linkage (string, optional) –
Linkage method of hierarchical clustering, see linkage for more details. The default is ‘ward’. Possible values are:
’single’.
’complete’.
’average’.
’weighted’.
’centroid’.
’median’.
’ward’.
’DBHT’. Direct Bubble Hierarchical Tree.
k (int, optional) – Number of clusters. This value is took instead of the optimal number of clusters calculated with the two difference gap statistic. The default is None.
max_k (int, optional) – Max number of clusters used by the two difference gap statistic to find the optimal number of clusters. The default is 10.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
leaf_order (bool, optional) – Indicates if the cluster are ordered so that the distance between successive leaves is minimal. The default is True.
- Returns:
A_c – Adjacency matrix of clusters.
- Return type:
ndarray
- Raises:
ValueError when the value cannot be calculated. –
Examples
C_M = rp.clusters_matrix(returns, codependence='pearson', linkage='ward', k=None, max_k=10)
The clusters matrix looks like this:
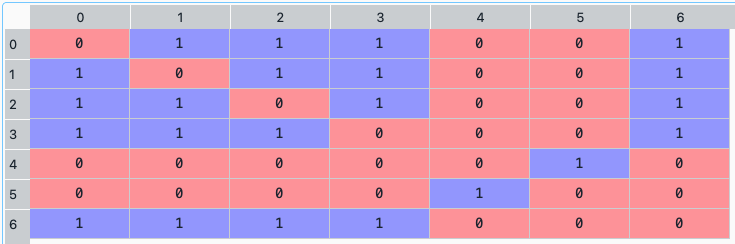
- ConstraintsFunctions.connected_assets(returns, w, custom_cov=None, codependence='pearson', graph='MST', walk_size=1, bins_info='KN', alpha_tail=0.05, gs_threshold=0.5)[source]
Calculates the percentage invested in connected assets based on [] formula.
- Parameters:
returns (DataFrame) – Assets returns.
w (DataFrame of shape (n_assets, 1)) – Portfolio weights.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
graph (string, optional) –
Graph used to build the adjacency matrix. The default is ‘MST’. Possible values are:
’MST’: Minimum Spanning Tree.
’TMFG’: Plannar Maximally Filtered Graph.
walk_size (int, optional) – Size of the walk represented by the adjacency matrix. The default is 1.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
- Returns:
CA – Percentage invested in connected assets.
- Return type:
- Raises:
ValueError when the value cannot be calculated. –
Examples
ca = rp.connected_assets(returns, w, codependence="pearson", graph="MST", walk_size=1)
Calculates the percentage invested in related assets based on [] formula.
- Parameters:
returns (DataFrame) – Assets returns.
w (DataFrame of shape (n_assets, 1)) – Portfolio weights.
custom_cov (DataFrame or None, optional) – Custom covariance matrix, used when codependence parameter has value ‘custom_cov’. The default is None.
codependence (str, can be {'pearson', 'spearman', 'abs_pearson', 'abs_spearman', 'distance', 'mutual_info', 'tail' or 'custom_cov'}) –
The codependence or similarity matrix used to build the distance metric and clusters. The default is ‘pearson’. Possible values are:
’pearson’: pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
’spearman’: spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{spearman}_{i,j})}\).
’kendall’: kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{kendall}_{i,j})}\).
’gerber1’: Gerber statistic 1 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber1}_{i,j})}\).
’gerber2’: Gerber statistic 2 correlation matrix. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{gerber2}_{i,j})}\).
’abs_pearson’: absolute value pearson correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_spearman’: absolute value spearman correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho_{i,j}|)}\).
’abs_kendall’: absolute value kendall correlation matrix. Distance formula: \(D_{i,j} = \sqrt{(1-|\rho^{kendall}_{i,j}|)}\).
’distance’: distance correlation matrix. Distance formula \(D_{i,j} = \sqrt{(1-\rho^{distance}_{i,j})}\).
’mutual_info’: mutual information matrix. Distance used is variation information matrix.
’tail’: lower tail dependence index matrix. Dissimilarity formula \(D_{i,j} = -\log{\lambda_{i,j}}\).
’custom_cov’: use custom correlation matrix based on the custom_cov parameter. Distance formula: \(D_{i,j} = \sqrt{0.5(1-\rho^{pearson}_{i,j})}\).
linkage (string, optional) –
Linkage method of hierarchical clustering, see linkage for more details. The default is ‘ward’. Possible values are:
’single’.
’complete’.
’average’.
’weighted’.
’centroid’.
’median’.
’ward’.
’DBHT’. Direct Bubble Hierarchical Tree.
k (int, optional) – Number of clusters. This value is took instead of the optimal number of clusters calculated with the two difference gap statistic. The default is None.
max_k (int, optional) – Max number of clusters used by the two difference gap statistic to find the optimal number of clusters. The default is 10.
Number of bins used to calculate variation of information. The default value is ‘KN’. Possible values are:
’KN’: Knuth’s choice method. See more in knuth_bin_width.
’FD’: Freedman–Diaconis’ choice method. See more in freedman_bin_width.
’SC’: Scotts’ choice method. See more in scott_bin_width.
’HGR’: Hacine-Gharbi and Ravier’ choice method.
int: integer value choice by user.
alpha_tail (float, optional) – Significance level for lower tail dependence index. The default is 0.05.
gs_threshold (float, optional) – Gerber statistic threshold. The default is 0.5.
leaf_order (bool, optional) – Indicates if the cluster are ordered so that the distance between successive leaves is minimal. The default is True.
- Returns:
RA – Percentage invested in related assets.
- Return type:
- Raises:
ValueError when the value cannot be calculated. –
Examples
ra = rp.related_assets(returns, w, codependence="pearson", linkage="ward", k=None, max_k=10)